This project is a tribute to my work as a molecular biologist. I have spent many years at the laboratory bench and the polymerase chain reaction (PCR) was one of my daily tasks. If you have ever done PCR, you probably know that it’s success, i.e. having a product or not, depends on many different parameters, such as primer design, template quality and reaction set-up.
Pipetting PCR master mixes can be a tedious task, especially if one has to look up every time after the optimal conditions in the product data sheet. Some of my colleagues used Excel worksheets for that purpose, but depending on how the worksheet is arranged, it is difficult to keep track of all parameters.
Therefore, I programmed a small app to calculate PCR master mixes, containing “hard coded” dilution factors and conditions for all kinds of polymerases available in my laboratory. This app turned out to be very useful, because it prevents calculation errors and helps not to forget important compounds for the master mix.
Programming a PCR-Calculator App with
GFA-BASIC
Back in the eighties, I did my first programming on a Commodore and used GFA-BASIC, developed by a German company (GFA Systemtechnik GmbH) as a dialect of the BASIC language. It was very popular on the Atari ST and was later even available for Windows. By the standards of its time, it was a very modern language and included even a GUI editor.
Although the GFA Systemtechnik GmbH doesn’t exist anymore, GFA-BASIC I discovered that is is still maintained and available for Windows (It can be downloaded from this page).
Therefore, I started this little fun project to revive good old memories …
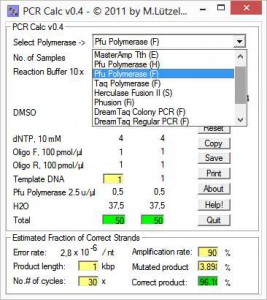
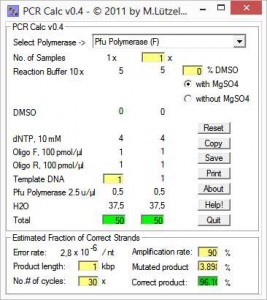
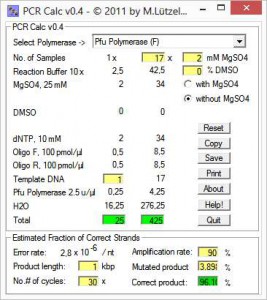
From a list, first you chose a polymerase going to be used for the PCR setup. Next, you enter number of reactions and reaction scale, e.g. 10, 20, 25, 50 or 100 μl. Special conditions, e.g. additional MgSO4 are activated upon polymerase selection.
All fields are refreshed as soon as you enter new values. When you’re done, you can estimate the probability for mutations in the PCR product using the lower panel. This is particularly useful, if you want to clone the product into an expression vector.